
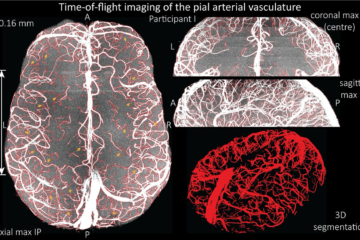
For the ISMRM 2016 we build a minimum deformation average (MDA) from a population of subjects based upon high resolution 7T MR imaging.
View online and download the model in MINC or NiFTI format here:
- View model online via tissuestack.org
- Download mnc (171 MB)
- Download nii (195 MB)
- code: https://github.com/CAISR/volgenmodel-nipype
Acquisition details
28 participants (21-34 years of age, 26.5 years on average, 14 males) on a 7 T whole-body research scanner (Siemens Healthcare, Erlangen, Germany), with maximum gradient strength of 70 mT/m and a slew rate of 200 mT/m/s. A 7 T Tx/32 channel Rx head array (Nova Medical, Wilmington, MA, USA) was used for radio frequency transmission and signal reception. We acquired data using a prototype ultra-short-TE sequence PETRA (Grodzki DM, Jakob PM, Heismann B. Ultrashort echo time imaging using pointwise encoding time reduction with radial acquisition (PETRA). Magn. Reson. Med. 2012;67:510–518. doi: 10.1002/mrm.23017.): TR = 1.99 ms, TE = 0.07 ms, flip angle = 2°, FOV = 288x288x288 mm3, matrix = 288x288x288 (1 mm isometric voxels), no GRAPPA, TA = 2 min. The volgenmodel pipeline was used to construct a minimum deformation model.First, the initial model was generated based on one individual dataset blurred using a kernel size of 4 mm to remove individual features. Then all original input images were aligned via a 12 parameter affine transformation and a normalized cross correlation objective function. The original input datasets were then resampled to the model space and transformed using a concatenation of the inverse transformation from model to participant space and the average transform. Finally, the next model stage was computed by using a robust averaging process of the resampled data, including only data within two standard deviations of the existing mean. After the affine transformation, non-linear fitting was used with incrementally decreasing step and smoothing kernel sizes.

0 Comments